This weekend I had the opportunity to participate in Def Hacks Virtual 2020, an online hackathon with tons of prizes and interesting workshops. The whole experience was incredibly fun, with thousands of participants from around the world and hundreds of interesting projects developed.
I am excited to announce that our project Synplifier won the Grand Prize at Def Hacks, receiving a grant of 1k cash and 10k in AWS credits from the 1517 Fund. I'm grateful for the support we received from the organizing team-- as primarily a college hackathon, the competition was definitely very tough. After developing the proof-of-concept over these past two days, I can see a lot of potential in it and will be continuing to develop it. My work was heavily inspired by my experiences on the SoundBio iGEM team last year, and also by the COVID-19 community project at the SoundBio Lab.
This post is mainly taken from the Devpost project, which you can view here as well. The code is accessible on Github, and you can demo this very early version yourself at my teammate's Pages. My teammate Ujjaini, also a rising senior, was incredibly helpful throughout the hackathon. I couldn't have done it without her.
As a way to grow diagnostic reagents from anywhere around the world, synthetic biology should allow for a proliferation of DIY viral testing. Yet the potential of synthetic biology remains locked behind complex lab protocols and largely restricted to highly specialized research fields. While the key to its potential, small DNA pieces known as plasmids, can easily travel around the world, the knowledge required to use them fails to do the same.
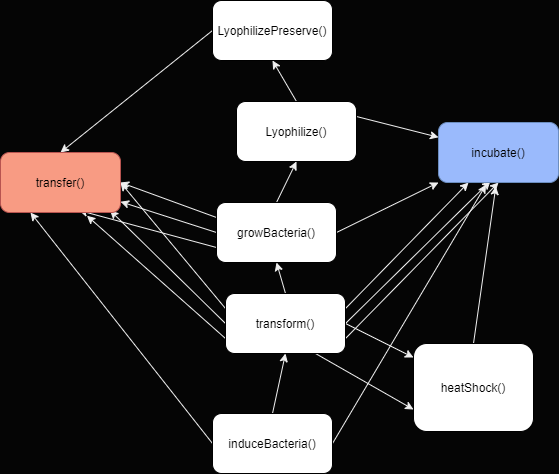
Synplifier allows scientists to translate production protocols into robust and user friendly walkthroughs. Think of it as a compiler for synthetic biology protocols-- going first from an academic lab protocol to standardized commands, and from standardized commands to a step-by-step breakdown. This simplification process relies on the modular nature of synthetic biology protocols. Essential protocols in synthetic biology-- growing, transforming, and inducing bacteria--are not difficult operations. At the most granular level, much can be reduced to the simple transfer of liquids or raising and lowering temperatures.

To demonstrate just how accessible synthetic biology can be, we took a COVID diagnostic protocol and wrote it into our pseudo compiler, a javascript p5js application. The result is a series of 86 simple commands (think “move 5ml of liquid from red cup to blue flask”) that can be followed without scientific literacy, fine motor control, or even knowledge of the English language. When developed in tandem with the test production kits, synplifier would be instrumental in the creation of test production capabilities anywhere in the world. In doing so, Synplifier and Synbio approaches to diagnostics aim to improve the lives of many--not through one time test distribution, but by investing in long term production.
The core functionality was developed in Javascript. The visualization and a prototype development environment are hosted on a static HTML page, with heavy use of P5.js for agile visualization.
One challenging part of the experience was integrating the logic into an effective visualization. Our initial goal was to create a scaffold for building AR walkthroughs in EchoAR, but we ultimately pulled away from its exciting possibilities to focus on the core product-- the simplification process itself.
By developing this project, we were able to dive into the construction of knowledge systems and human computer interaction. We also obtained a greater understanding of synthetic biology and LAMP-DETECTR procedures through writing functions to formalize their specifications. And of course, we also learned about styling pages and libraries such as Bootstrap.
We see Synplifier as a very strong proof of concept for an LIMS with a unique focus. As of now, Synplifier is effective in providing archetypal 2-dimensional visualizations of lab procedures, with a limited range of flexibility. We hope to increase its functionalities greatly moving forward and get user feedback from both the scientific community and other interested parties. We also hope to look further into distributed diagnostics and how Synplifier might work in conjunction with labs to reach vulnerable and at-risk communities.